In vitro retinal disease modelling for retinal therapy
Newcells offers a fast and reliable in vitro safety and efficacy service for the evaluation of novel compounds for retinal therapy using complex human retinal organoid or RPE models developed in house. Both models are iPSC derived using WT line or CRISPR/Cas9 gene edited lines supplied by our customer for direct comparison between WT and mutant phenotypes after differentiation. Targeted mutations can model retinopathies, more specifically monogenic inherited retinopathies such as retinitis pigmentosa (RP), Stargardt disease, Usher’s syndrome and Leber congenital amaurosis.
Service outputs
- Cell viability assay
- Key markers analysis
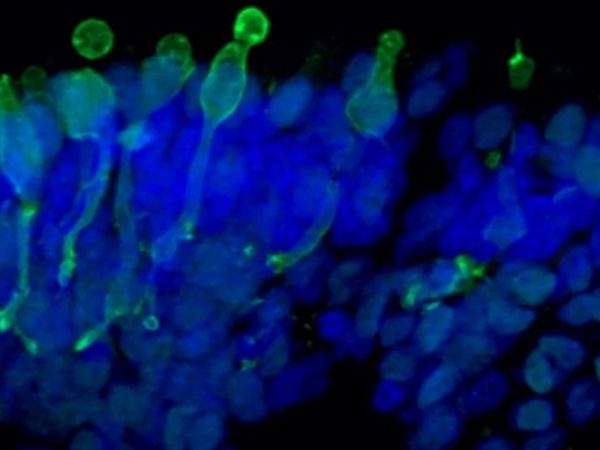
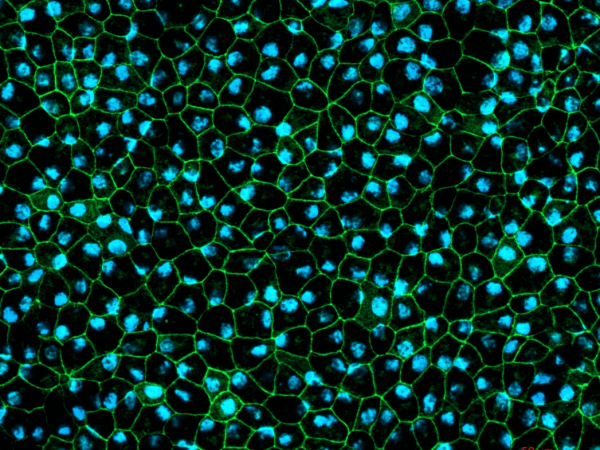
- Qualitative immunofluorescence
- Gene expression
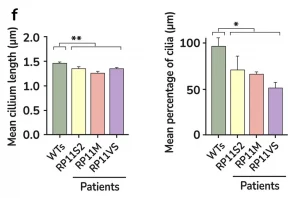
- Photoreceptor degeneration
- Phagocytosis of photoreceptor outer segment (RPE)
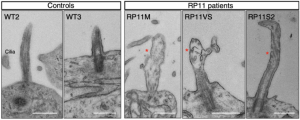
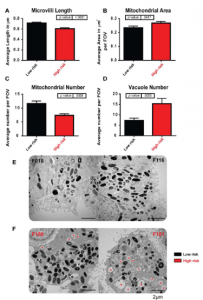
- Transmission (TEM) and Scanning (SEM) electron microscopy